Contact Usss
- Tong Hao for questions regarding the database
- David Hill for questions regarding the ORFeome project
- Horizon Discovery
Promoter Collection
for requesting any ORF clones - Promoterome Database
for promoters
Tech Support
Please email Tong Hao with your questions or comments.
Notice:
This website does not support IE 5 or earlier IE version. Please use a updated web browser.
Liability:
For
documents and software available from this server, there is not warrant
or assume any legal liability or responsibility for the accuracy,
completeness, or usefulness of any information, apparatus, product, or
process disclosed.
About The RACE Project
The C. elegans genome has been fully sequenced, but experimental verification of over a third of the predicted genes remains outstanding. Though ORF predictions are better for the C. elegans genome than for other metazoans, still ORFs for ~8,000 predicted genes remain experimentally unverified in part because of problems with current exon/intron structure predictions. We have undertaken to experimentally define transcript structures of a large portion of these unverified genes by RACE (Rapid Amplification of cDNA Ends), so far we have carried out 5' and 3' RACE reactions for ~2,000 of the ~8,000 unverified C. elegans protein coding genes, which are included in the searchable database available in this page. These data can also be accessed through WormBase .
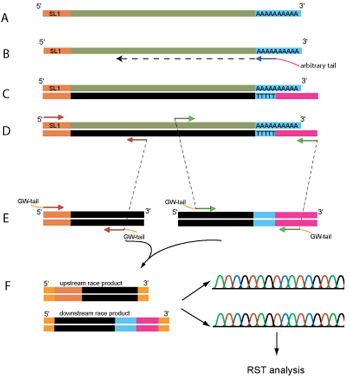
General schema for 5' and 3' RACE experiments. Messenger RNA (A) is reverse transcribed with a tailed dTn primer (B) to generate complementary DNA strand (C), used as template (D) to generate the first sets of RACE products (E). These amplicons are then reamplified with the nested internal primers to generate the final RACE products (F), which are cloned, sequenced, and analyzed.
The general scheme for our RACE experiments is shown in the figure. For our 5' RACE experiments we made use of the trans-spliced SL1 and SL2 leader sequences, instead of ligating a universal sequence to 5' of the transcripts. Approximately 70% of all C. elegans mRNAs have a trans-spliced leader sequence. The great majority of these correspond to a 22 base-long sequence known as "SL1," with "SL2" being the next most frequent, making up 15% of the worm's trans-spliced leaders. The use of SL1/SL2, as opposed to the ligation of an arbitrary sequence to the transcripts' 5' ends has the following advantages: i) no additional manipulation of RNA is needed, ii) the presence of SL1 on a mRNA ensures that the mRNA has an intact 5' end and is full length.
To generate the RACE fragments, we reverse transcribed total C. elegans RNA (isolated from mix-stage, asynchronously growing N2 worm population) using either dT16 (for 5' RACE) or used our tailed dT primer (for 3' RACE). Nested PCRs were performed to increase sensitivity and specificity. The generated PCR products were then cloned recombinationally and sequenced from the 5' end, generating "RACE Sequence Tags" (or "RSTs").
The RSTs included in this searchable database are vector and quality trimmed (SL and poly(A) sequences were not removed from RSTs). In quality trimming, the first sliding window of 20 nt long with an average quality score higher than 15 marks the start of good quality sequences. Likewise, the first sliding window of 20 nt with average quality score lower than 15 marks the end of good quality sequences.